Note: This writeup ended up being 10x longer than we expected. A few have already asked for a PDF or downloadable version. We are working on formatting a portable version of this document for offline reading and distribution. Leave a comment or drop us a line if you want an offline version.
Intro
One of the great things about working for the BioTeam is that we get to work with (and within!) many different organizations each year. It’s an impressively diverse set of groups as well — from incubation stage startups still operating in stealth mode through global pharmaceutical companies. Outside of the commercial space we also have a significant client base comprising nonprofit research institutes, academic institutions and US Government labs and agencies.
All this consulting work allows us to see the real world, in-the-trenches view of how many different groups of smart people approach “Bio-IT” or life science informatics problems and challenges.
Unfortunately we are often too busy to formally organize and distribute information about the trends we see, the lessons we’ve learned and the problems we continue to face.  Our website also tends to lag very far behind in describing and talking about what we are currently working on. Many people have noted that our most up-to-date advice, observations and tips tend to be delivered verbally when we speak at industry conferences, workshops and technical events. We do have a good track record of making our presentation and talk slides available online but PowerPoint can be a poor medium for conveying information outside of a live presentation or event. Due to popular demand, this post is being written in an attempt to distill many of our storage and NGS data management related presentations into a written form that can be more easily be updated, discussed and distributed. So – if you’ve seen BioTeam public talks such as:
Our website also tends to lag very far behind in describing and talking about what we are currently working on. Many people have noted that our most up-to-date advice, observations and tips tend to be delivered verbally when we speak at industry conferences, workshops and technical events. We do have a good track record of making our presentation and talk slides available online but PowerPoint can be a poor medium for conveying information outside of a live presentation or event. Due to popular demand, this post is being written in an attempt to distill many of our storage and NGS data management related presentations into a written form that can be more easily be updated, discussed and distributed. So – if you’ve seen BioTeam public talks such as:
… this blog post will not contain many surprises as it will generally recap and put in writing the things we’ve been saying in front of audiences for quite some time. However, for those that have not attended our talks, please consider this post as our attempt to “brain dump” into written form our current perspective on the life science storage, NGS and data management landscape. As always – comments, corrections and feedback always appreciated!
Meta: The core problem influencing all Bio-IT storage and data management efforts
1. Scientific innovation rapidly outpacing IT innovation
It’s a fairly scary and potentially risky time to be working on IT systems designed for use in life science environments. The simple truth over the last few years is that the technical requirements and technologies in use by today’s scientists change far faster than even the best funded and most aggressive organizations can refresh or alter their IT infrastructures. 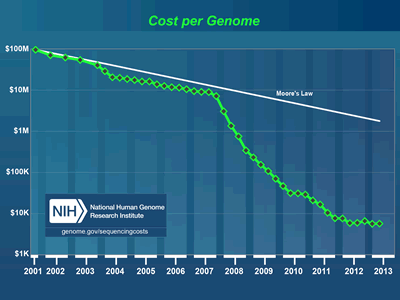 The rate of change and innovation in the lab is outpacing even Moore’s Law. A great example of this in chart form can be seen over at the NIH Sequencing Cost page: http://www.genome.gov/sequencingcosts/ showing just how quickly the costs of genomic sequencing have fallen over the past few years. We somehow have to deal with the fact that major algorithm, instrument and laboratory SOP changes are happening almost monthly and that each time a new innovation, tool or sensor is deployed it can have a very significant and immediate impact on IT infrastructure. BioTeam’s best funded and most aggressive clients can typically afford a planned IT refresh every two years or so. Our larger enterprise clients typically operate scientific and research IT platforms over a 3-5 year lifespan before major refresh or retirement efforts are budgeted for. Add in the fact that major facility upgrades (electrical and cooling) or new datacenter construction can take years to plan and implement and we have an incredibly scary research IT landscape to navigate.
The rate of change and innovation in the lab is outpacing even Moore’s Law. A great example of this in chart form can be seen over at the NIH Sequencing Cost page: http://www.genome.gov/sequencingcosts/ showing just how quickly the costs of genomic sequencing have fallen over the past few years. We somehow have to deal with the fact that major algorithm, instrument and laboratory SOP changes are happening almost monthly and that each time a new innovation, tool or sensor is deployed it can have a very significant and immediate impact on IT infrastructure. BioTeam’s best funded and most aggressive clients can typically afford a planned IT refresh every two years or so. Our larger enterprise clients typically operate scientific and research IT platforms over a 3-5 year lifespan before major refresh or retirement efforts are budgeted for. Add in the fact that major facility upgrades (electrical and cooling) or new datacenter construction can take years to plan and implement and we have an incredibly scary research IT landscape to navigate.
In a nutshell, today’s Bio-IT professionals have to design, deploy and support IT infrastructures with life cycles measured over several years in the face of an innovation explosion where major laboratory and research enhancements arrive on the scene every few months.
2. IT often the last to know about major lab-side changes
One of the recurring themes encountered out in the real world is how IT organizations are often taken utterly by surprise when new laboratory instruments or techniques arrive on-premise and immediately require non-trivial amounts of IT resources. Here are just a few examples we’ve seen over the past few years:
- Instrument Upgrades: In place upgrades to existing instruments can often slip under the radar of even the most watchful IT organizations. The cliché example here would be the Illumina HiSeq genome sequencing platform where a HiSeq 2000 instrument can be upgraded to a HiSeq 2500 by swapping flow cells and reagents. The IT requirements for a HiSeq 2500 can be quite a bit higher.
- Instrument Duty Cycle Changes: IT resources are often provisioned for instruments based on an understanding of the common duty cycle. Often the first use of an instrument is for basic experimentation and validation of the intended protocol and result output. When the results are good, scientific leadership may decide to dramatically change the way the instrument is used. The resources required for an instrument that runs for a few hours a week followed by two weeks of data processing is quite different from an instrument that is operated and scheduled 24×7 in a core facility operational model.
- New Sensors: A scientist took the “regular” camera off of the confocal microscope rig and replaced it with a new CCD sensor capable of capturing 15,000 video frames per second. IT was not informed and the microscope storage platform was not altered.
- DIY Innovation: A scientist had trouble using a confocal microscope for live cell imaging experiments — the cells being examined did not survive long under the microscope. Working with a few colleagues they hacked together a clever DIY incubation enclosure around the microscope rig to better control environmental conditions. All of a sudden live cell imaging efforts that previously could only last for 20-40 minutes are being run for 24-hour or even longer periods. Demand for storage, compute and visualization resources spikes accordingly.
- Broken Procurement: In general terms this is what happens when researchers spend 100% of their budget on the instrument and the reagent kits (and perhaps an operator to run the machine as well) while neglecting to plan or budget for the IT resources needed to sustain operation and downstream analysis. This problem used to be much worse in years past where we saw instrument salespeople outright lying to customers about IT requirements and cost in order to win a sale. In 2013 and beyond we still continue to see poorly-managed laboratory instrument procurement processes. Given the data flows coming from these instruments it is essential that procurement is able to model, plan and budget for the full lifecycle cost of the instrument. This includes instrument data capture, QC efforts, data movement, data storage, processing/analytical resources as well as long-term or archival storage of both the raw and derived data.
3. The “easy” period is over


The “easy” period ended several years ago when the scale of the problem began to overwhelm the cheap, simple and local techniques professionals in our field had been using.
Most of these techniques involved installing IT hardware “close” to the lab — often in the form of tower form-factor server systems or small cabinets that could fit under a wet lab bench or next to a desk.
Another very common method involved guerrilla takeovers of nearby available telco or communication closets used to provide network and telephone connections to everyone on the building floor. Many small disk arrays, servers and even mini Linux compute clusters started showing up in data closets and communications rooms proximate to the lab spaces where high-rate data producing instruments were operating.
For several years now, the older methods have been falling out of favor. The scale of the problem is large enough that systems no longer fit conveniently under a desk or in a nearby telecom room. When a single instrument may require 60 terabytes of disk and a few dozen CPU cores just to handle daily ingest and operation (ignoring the additional disk and CPU requirements for downstream analysis) it is clear that datacenter class solutions and systems are required. Today the only major deployment of lab side IT hardware is primarily for supporting instrument control workstations and (frequently) caching instrument output to a local storage array so the technician can run final QC/QA tests before allowing the data to move on into the analysis and archive environment.
In 2013 our bare-minimum Bio-IT footprint often starts out at “hundreds of cores” and “peta-capable storage” — both of which are difficult (or risky) to deploy anywhere other than a dedicated datacenter. It is not uncommon for us to be working on infrastructures where 1000 CPU cores and 1000 terabytes of raw disk are operating in support of less than 40 researchers. For sure, the “easy” period has long been over!
4. It has never before been easier for scientists to generate or acquire vast amounts of data
In a nutshell: The rate at which scientists can generate or acquire data exceeds the rate at which the storage industry is making disk drives larger

When examining the number and variety of “terabyte capable” lab instruments combined with the massive amount of biological information available for download via the internet (Examples: NCBI, 1000 Genomes Project, UK10k, Amazon Public Data Repositories) it quickly becomes apparent that we are in an unprecedented era for the life sciences — the sheer amount and variety of data that can be generated or easily acquired by a single researcher or small group of scientists can very quickly swamp even the largest enterprise storage platforms.
And this is just the baseline or “input” data! We still need additional compute and storage resources to handle the actual science being done on this data and all of the new files and output produced.
Bio-IT insiders who have been charting the growth of file and data usage on internal systems can see the trend clearly — the increasing rate at which scientists are accumulating data seemingly exceeds the rate at which the storage industry is making disk drive platters denser. This is very worrying to an IT professional trying to plan several years ahead into the future.
How the ‘meta’ issues affect storage and data management efforts today
In the prior sections, I’ve tried to describe some of the background and “meta” issues on the minds of many people responsible for research computing and storage infrastructures in the life sciences. Here is a listing of how those concerns and issues often affect real-world storage and infrastructure projects in our field:
-
- Capacity over Performance:
 Selling storage on raw performance alone is a losing proposition for a storage vendor. Life science customers are worried about explosive data growth and being able to handle the high rates of innovations coming out of the laboratory environments. The end result is that they are consciously choosing to deploy systems that may offer slower raw performance but compensate by offering massive scalability or a much lower dollar cost per terabyte. The future is so uncertain and hard to plan for in discovery-oriented R&D environments that conscious decisions are being made to favor systems with a better chance of surviving a multi-year lifecycle where the primary requirement is the ability to support explosive growth and unpredictable novel use cases. In practical terms this is why I’ve personally built and deployed many EMC Isilon scale-out storage clusters that use NL-series nodes as primary building blocks. The NL product line messaging often includes the words “archive” or “nearline” but the blunt truth is that these are used very often in life science environments where the cost per terabyte ratio is of high importance and the ‘faster’ Isilon nodes types are often cost prohibitive for the general business case.
Selling storage on raw performance alone is a losing proposition for a storage vendor. Life science customers are worried about explosive data growth and being able to handle the high rates of innovations coming out of the laboratory environments. The end result is that they are consciously choosing to deploy systems that may offer slower raw performance but compensate by offering massive scalability or a much lower dollar cost per terabyte. The future is so uncertain and hard to plan for in discovery-oriented R&D environments that conscious decisions are being made to favor systems with a better chance of surviving a multi-year lifecycle where the primary requirement is the ability to support explosive growth and unpredictable novel use cases. In practical terms this is why I’ve personally built and deployed many EMC Isilon scale-out storage clusters that use NL-series nodes as primary building blocks. The NL product line messaging often includes the words “archive” or “nearline” but the blunt truth is that these are used very often in life science environments where the cost per terabyte ratio is of high importance and the ‘faster’ Isilon nodes types are often cost prohibitive for the general business case.
-
- Operational Burden Matters: I (and many others) will pay a higher price for IT systems that deliver clear and tangible benefits when viewed from a management and operational burden viewpoint. A system that takes less resources to run and manage is more desirable. Assessment of operational burden may become the deciding factor in picking a winning bid from a vendor. Getting approval for capital expenditures is easier than getting approval to hire additional human hands! Outside of academic environments where grad students and postdocs are leveraged as labor sources it is common now for new IT systems (especially storage) to be evaluated specifically on how complex the system is and how difficult (or easy) it is to manage and maintain on a daily basis. There is a famous Bio-IT “war story” from years ago at the Bio IT World Expo & Conference where a senior Research/IT leader from a very well-known research institution with many petabytes of data under management stood at the podium and related a tale that basically boiled down to this: he had a staff of several (half-dozen?) storage engineers working almost full time to keep the inexpensive Sun Thumper arrays from falling over while he, himself, was able to manage a 6-petabyte Isilon cluster in his spare time. That particular Bio-IT talk and the reverberations that came from it as his story spread had a huge impact on our field. It really reinforced the message that “cheap storage may not be so cheap” if it requires non-trivial human resources to keep running. For many years Isilon sold a ton of systems into the life sciences and ran rings around competitors because their sales messaging concentrated on scalability and ease-of-management while others were still touting raw IOPS/second figures as the primary reason for consideration.
-
- Simplicity (may) Matter: System complexity and operational burden issues can influence storage design patterns as well. This is why I tend to see many more scale-out NAS systems in our space instead of large SAN-based or distributed/parallel filesystems running GPFS or LUSTRE. There are many ways to deploy large storage but grafting one or more filer gateways onto a SAN or dealing with all of the complex compute, storage, interconnect and metadata controller issues found in modern parallel filesystems can be a challenge. We love big and fast parallel filesystems as much as anyone but we’ve seen enough at client sites to take note of a clear pattern. There are two types of successful organizations with deployed parallel filesystems: those that invest in full-time onsite specialist expertise and those that purchase a fully-supported stack (hardware, software, client connectivity, etc.) from vendors such as DataDirect who will be on the hook for everything from pre-purchase design to ongoing support.
In the face of explosive current growth and research innovations that regularly outpace the rate at which IT can refresh infrastructures and datacenters, today’s Bio-IT organizations are attempting to mitigate risk and deal with unknown futures with as much agility as possible. These groups are gravitating towards petabyte-capable (because they can’t forecast much beyond 8-10 months), scale-out (because they can’t forecast rate of growth) network attached storage (NAS) systems (NAS is the best general fit for use cases that demand simultaneous multi-protocol, multi-client access to shared data). These organizations will trade down on speed and performance if it will gain them significant capacity or agility and they will seriously evaluate potential solutions in order to characterize ongoing operational and administrative burden.
What Bio-IT Organizations Are Doing Differently Today
-
- Science/IT Outreach and Expectation Setting: Many years ago at the start of my career, an IT “ah-ha!” moment occurred during a discussion with some scientists concerning a short term need for immediate storage. I was trying to explain to them why their request to add many dozens of terabytes of capacity to an existing SAN was technically complicated and would be incredibly expensive. When trying to understand why they reacted with a bit of hostility and some subtle hints that “IT is really bad at its job…” I realized that this group of researchers had viewed the cost of enterprise IT storage as being roughly comparable to what it would cost them to purchase the equivalent capacity in the form of external USB drives from places such as Staples, Amazon.com and Best Buy. With this internalized assumption about storage costs they were shocked and outraged when I told them the actual price of upgrading the corporate SAN to the capacity they were demanding. No wonder they thought I was really bad at my job! Once I had that realization I was able to turn the situation around and we had a great and cordial discussion concerning the true full-lifecycle costs of keeping 1 terabyte of scientific data safe, secure and always accessible within a single namespace. Using a whiteboard we discussed all of the systems, hardware, staffing and support contracts required to keep the data safe, backed-up, replicated and instantly accessible. I can’t remember the exact price we arrived at but this was a high-end enterprise-class block-storage SAN in the era before scale-out architectures and 1TB drive platters. I think we arrived at a rough cost of $10K-15K per terabyte. This was shocking to the scientists who had long adopted work and data management patterns based on an implicit assumption that storage was not a particularly scarce or expensive commodity. Once we had the “true cost of scientific storage” discussion with the scientists the impact was immediately visible – the researchers began changing their behavior and data management patterns and became active collaborative partners with the IT organization. I’ve now repeated this sort of discussion enough times over the years to fully understand just how much influence the “street price” of consumer-grade external storage can have on the behavior of scientists and non-IT professionals. Most people truly don’t know the real world costs associated with keeping massive amounts of data online, safe and available. This is no fault of their own but it is the responsibility of the IT organization to set proper expectations, have the proper conversations and actively collaborate with researchers. The most successful Bio-IT organizations perform outreach efforts and set expectations early and often.
-
- Data Triage: Data triage and deletion of scientific data is a mainstream practice across the entire organizational spectrum – even the oldest and most conservative pharmaceutical companies. It is clear now that for certain types of experiments, particularly genomic sequencing it is far cheaper to go back to the -40 F sample freezer and repeat the experiment than store all of the raw experimental data online, forever. The one exception we’ve seen is within some US Government labs where rigid information technology guidelines can have a paralytic effect on government employees who may be unwilling to delete any data generated off of any instrument without a written order and multiple approval signatures. The one important thing to note: it is never appropriate for IT staff to make a data deletion or triage decision on their own — all such decisions must come from the science side. This is another major reason why outreach and expectation setting needs to be done with research users — few scientists will entertain a request to delete scientific data if they’ve never had the “true cost of scientific storage” talk.
-
- Traditional backup methods becoming scarce:
 Traditional tape-based backup is now incredibly rare in our space – not used at all or perhaps only used to preserve a very tiny percentage of data under management. Other enterprise practices like high-end enterprise storage replication between matched systems is falling out of favor as storage grows above and beyond the 200-300 terabyte raw range and the cost of the replica systems make a big dent in budgets. Many of our clients are simply not taking extraordinary measures to backup or replicate data, having done a risk assessment and deciding to live with the risks of operating out of a single site and facility. Others are starting to take more labor intensive but far less expensive DIY-style efforts such as using rsync to mirror data between dissimilar systems such as an EMC Isilon with homegrown data replication flowing to far denser and less expensive storage systems by vendors such as Nexsan or disk-heavy ZFS-based systems from companies like Silicon Mechanics or systems running Nexenta software. In small genomic environments where data volumes are small enough (under a few tens of terabytes) we also have heard of people replicating to multiple Drobo units and simply taking the Drobo’s home or swapping them occasionally in an offsite location. Others use independent systems that may offer data migration or movement as a feature such as Avere FXT nodes. The Avere systems are primarily used for storage acceleration and caching but they can also consolidate different systems under a single namespace as well as move data between different storage tiers or even Amazon S3 and Glacier.
Traditional tape-based backup is now incredibly rare in our space – not used at all or perhaps only used to preserve a very tiny percentage of data under management. Other enterprise practices like high-end enterprise storage replication between matched systems is falling out of favor as storage grows above and beyond the 200-300 terabyte raw range and the cost of the replica systems make a big dent in budgets. Many of our clients are simply not taking extraordinary measures to backup or replicate data, having done a risk assessment and deciding to live with the risks of operating out of a single site and facility. Others are starting to take more labor intensive but far less expensive DIY-style efforts such as using rsync to mirror data between dissimilar systems such as an EMC Isilon with homegrown data replication flowing to far denser and less expensive storage systems by vendors such as Nexsan or disk-heavy ZFS-based systems from companies like Silicon Mechanics or systems running Nexenta software. In small genomic environments where data volumes are small enough (under a few tens of terabytes) we also have heard of people replicating to multiple Drobo units and simply taking the Drobo’s home or swapping them occasionally in an offsite location. Others use independent systems that may offer data migration or movement as a feature such as Avere FXT nodes. The Avere systems are primarily used for storage acceleration and caching but they can also consolidate different systems under a single namespace as well as move data between different storage tiers or even Amazon S3 and Glacier.
- Traditional backup methods becoming scarce:
-
- New Roles and Responsibilities: More and more clients of ours are opening up new hybrid Science/IT jobs with responsibilities that can be roughly described as “Data Manager” or “Data Curator”. The cost of storage systems required to support the explosive growth in scientific data under management is now high enough that the associated cost of hiring a human being to assist with data classification, tiering, archiving and policy decisions is seen as a relative bargain. People are operating at scales today where a huge amount of information (and potential cost savings) are flying under the radar because the resources do not exist to maintain a careful eye on what (and where!) things are being stored.
- Show back and Chargeback: The classical argument against storage chargeback and show back models was that the effort and/or cost required to implement them properly was high enough that it outweighed the benefits. I don’t think this is the case any longer. The more visibility the better. It is also abundantly clear that transparent IT consumption metrics published within an organization have an undeniable effect on usage patterns and behaviors. Even a simple monthly list of “Top Ten Storage Users” can have a peer pressure effect that influences researchers to be more careful about the resources being consumed. Nobody wants to be on the “Top Ten” list without a valid scientific requirement. Chargeback is something that I philosophically have an issue with and have never really implemented myself but the anecdotes from the field are starting to become more common. One great heard-from-a-friend anecdote comes from the Broad Institute where a show back storage model had long been used. When the Broad Institute decided to switch from show back to a chargeback model there was a period during the 30-days before the new chargeback policy went live when 1 petabyte of disk suddenly became free and available. Think about that for a moment — simply by moving to a chargeback policy this particular organization suddenly saw 1 petabyte of storage disappear from the system resulting in more than $1M dollars in capital hardware becoming available for use. Amazing.
Life Science “Data Deluge” – Reasons not to panic
I sometimes get tired of breathless marketing aimed at our segment of the market, here are a few arguments I used to rebut people trying to scare the heck out of my friends, colleagues and clients:
Most of us are not Broad, BGI or Sanger
There is a small group of “extreme scale” life science informatics environments such as the Broad Institute, BGI or the Sanger Institute who operate daily at the extreme end of the Bio-IT spectrum. These folks routinely run multi-megawatt datacenters, consume tens of thousands of CPU cores and have dozens of petabytes of scientific data under management. At this scale it is not uncommon to see hundreds of terabytes of storage deployed each month. The scale at which these entities operate is different enough that they look, feel and operationally behave quite differently from “the rest of us”. They employ dedicated technical staff with deep expertise and may be operating exotic platforms and technologies more traditionally associated with supercomputing installations funded with sovereign nation resources.
Our needs are well within the bounds of what traditional “Enterprise IT” can handle
99% of us are not in the extreme tier of IT infrastructure consumers or operators. We may have petabytes of data and require thousands of CPU cores but the blunt truth is that in 2013 the vast majority of us operate solidly within the reasonable realm of the Enterprise IT envelope. Our needs are big and the solutions may be expensive but they are not particularly strange, exotic or unusual. The vast majority of us have IT issues that can easily be handled by standard enterprise scale-out architectures and well-understood methods and technologies.
Bio-IT practitioners have been doing this for years and the sky has not fallen
After a certain time the constant warnings of imminent doom tend to grow tiresome. Another blunt truth is that we’ve been dealing with “terabyte tsunami” scare stories since the first next-gen sequencing platforms came on the market in the early 2000’s. Sure it’s been a headache at times and we’ve seen a few disasters over the years but people are smart, adaptable and learn from past mistakes. We may not be operating at the speed, size or efficiency that we’d like but most of us have managed to muddle through and “keep the science going” in the face of every IT, storage or data handling problem that has come our way.
Instruments continue to get smarter
The days of ingesting terabyte volumes of raw .TIFF images off of DNA sequencers are over for most of us. Each new generation of instrument gets better and better at performing in-instrument data reduction and outputting only the actual scientific data we care about. The current generation of genomic sequencing platforms are able to spit out manageable streams of base calls and quality scores. Many of these instruments have reduced data flows to the point where previously infeasible methods like “write everything to remote storage” or even “write directly to the cloud” possible and practical.
Petascale storage no longer exotic nor risky
A decade ago, rolling out a petabyte-class storage system involved taking a leap of faith onto a set of products and technologies that usually could only be found in the largest of the global supercomputing sites. This was a risky endeavor involving smaller, niche technology vendors with potentially “career limiting” effects if things went sideways. They were expensive, hard to procure and needed deep expertise during pre-sale, post-sale, deployment and ongoing operation.
In 2013, petabyte-scale storage is no longer rare, exotic or risky. There are a dozen vendors who will happily deliver petabytes of fully-supported storage to your loading dock in a few weeks (or a few days if you catch them close to a fiscal year or quarter-end). It’s not a big deal any more — simply an engineering, budgeting and IT procurement exercise.
Life Science “Data Deluge” – Ok maybe panic a little …
After calming myself down with the points listed above, here are a few things that I do actually stress out about. Maybe a little panic is in order!
Those @!*#&^@ Scientists …
The pesky human scientists are responsible for wiping out all of the “gains” seen via instruments getting smarter about data reduction. The additional ‘derived’ data generated, manipulated and mashed-up by human researchers working with the instrument output is growing faster than the instruments themselves are reducing their overall data flows.
Humans are far harder to model and predict than instruments with known duty cycles. The end result is that it is now far harder to do capacity planning and storage prediction when the primary drivers for storage consumption have transitioned from instrument to researcher.
@!*#&^@ Scientific Leadership …
Lets revisit that “Cost of Sequencing” chart over at http://www.genome.gov/sequencingcosts/ — it is amazing to see the cost of sequencing fall faster than Moore’s Law. Do you think that translates into lower IT costs? Nope.
Sequencing is now a commodity. Nobody ever just banks the financial savings and keeps on working. If a genome sequencer is suddenly 50% cheaper, people will just buy two instruments and double the amount of work being done.
Current trends seem unsustainable …
Lets recap a few things, all of which when looked at as a whole can be pretty sobering:
- It has never before been so easy for life science researchers to generate or acquire immense volumes of data
- Increased rates of downstream storage consumption by human researchers has wiped out the gains seen by instruments doing better data reduction
- The intense commoditization curve affecting many “terabyte instruments” simply means people are purchasing more and more of them as they get cheaper
- Pace of innovation in the research lab now moving far faster than IT infrastructure can adapt
This confluence of events does not appear sustainable to me (and others). Something is going to break and we will either see radical changes or radical disruption in many research oriented life science environments.
Traditional Compression and Deduplication: Not good enough
The short story here is that if life science is going to continue at the same rapid rate of innovation including the ability to handle thousands or even tens of thousands of genomes for medicine, metagenomics or GWAS work etc. than incremental improvements (10%, 40%, 50%, etc. etc.) are simply not going to be sufficient.
We need to improve our ability to store and manage data by an order of magnitude.
Others make this argument better than I and Ewan Birney’s initial writings discussing the potential of reference-based CRAM compression for genomic data make for a great overview of the problem and one potential solution (CRAM-based compression) that is gaining traction. Check these articles out:
- Compressing DNA: The future plan
- Engineering around reference based compression
- Compressing DNA: Part 1
Technical Requirements for Life Science Storage
The overwhelming use case in research environments is the requirement for simultaneous, shared, read-write access to files and data between instruments producing data, HPC environments processing data and the research desktops used in visualizing and assessing the results. Everything else flows from this base requirements. This base requirement is also why scale-out NAS is the most popular implementation approach.
Must-have Features
-
- High capacity and scaling headroom: See above for why this is essential.
-
- Support for diverse file types and access patterns: We have hundreds of apps and dozens of file types and formats in common use. There is no standard file size, block size or access pattern.
-
- Multi-protocol access: Instrument control workstations and other inflexible clients with fixed and unchangeable network file access protocol requirements are common. CIFS and NFS usually.
-
- Simultaneous shared access: Multiple instruments, systems and people need shared access to the same file. A block storage SAN LUN exposed to a single host is insufficient.
- Post RAID-5 features: RAID5 is a disaster in large-disk/dense-array environments where a 2nd drive failure during a rebuild of a previous failed drive becomes a statistically more probable event. The specific method for surviving drive failures does not matter (RAID6 is sufficient along with many other methods) is not as critical as the storage platform having some reliable method of handling multiple concurrent drive failures.
Nice-to-have Features
-
- Large single-namespace support: This may belong in the “must-have” list but will stay here for now. There is a huge and compelling advantage to products supporting a large single namespace for files and folders. The evidence has been clear for many years – scientists given access to disconnected islands of storage are rarely able to manage them effectively. The end result is that data is stored on every island and issues of data provenance or “which file is the most current?” quickly appear. Being able to organize folders under a single namespace that can expand non-disruptively over time is a huge capability and efficiency win for Bio-IT organizations.
-
- Low operational burden: This has already been mentioned above. Inexpensive storage solutions are not a wise choice if they require hiring additional staff to keep them running well so procurement decisions based purely on price per terabyte are misguided. The price of a system needs to be evaluated in the context of the resources required to operate and maintain it.
-
- Appropriate pricing/market models: This is an aging complaint of ours dating back to the times when simply to get storage systems large enough to handle our capacity requirements we were often forced into the most expensive of the “Enterprise” storage product stacks larded up with all sorts of features, tricks and capabilities that we had to pay for and would never ever use. This complaint has largely subsided now that vendors are tuning product and price offerings for the “big storage without all the bells and whistles” crowd.
-
- Tiering Options: There have been a lot of monolithic scale-out NAS platforms deployed in the life science space over the last few years. For some storage products offered by well-known vendors, tiers have been a hard sell into the market because of the significant upfront investment required to fully buy-in to the additional tiers. This is why many of the infrastructures we see today are still operating off of the single “capacity optimized” that was originally purchased. Anything that allows or enables flexible tiered options for active or archival use is a positive.
-
- Inexpensive speed boost options: Our market has a lot of incumbent monolithic storage tiers built from slower capacity-optimized disks, nodes and shelves. Vendor products or features that allow modest amounts of additional performance to be injected into the environment without extraordinary cost can be very attractive. This is one of the reasons I like the Nexsan storage line – they sell scale-out NAS systems built via piles of 4TB SATA disks but you can still insert a couple of solid state SSD drives into most of their chassis for a quick (and invisible to the user) cache and performance enhancement. This is also where systems like Avere are generating interest as well, as their systems can be used to add a performance tier in front of a big pile of slower monolithic NAS storage, or can be used to front-end many different types of public or private clouds or on-premise storage tiers.
- Replication, movement and cloud storage options: Features that make moving and migrating data less of a hassle are always attractive. Cloud storage is simply a RESTful HTTP call away so modern storage platforms should be expected to treat remote object stores as simply another (offsite) storage tier or target. Modern storage controllers and systems almost all seem to run some sort of “Unix-like” environment under the hood. It’s a no-brainer that they continue to get smarter, more powerful and more capable.
Data Size, File Types and Access Patterns
Storage vendors, especially presale engineers are always deeply interested in details access patterns, typical file size and types. The bad news is that unlike other industries and vertical markets we do not have a small group of universally used files, sizes or standard formats.
The real answer is that discovery oriented life science environments have a little bit of everything. Sales people used to waxing poetic at how well their product handles a particular issue (very small files for instances) will need to broaden their messaging to speak about how their products perform across the entire spectrum.
A few examples:
-
- Many tiny files in a single directory: This situation is notorious for tripping up storage products in which inode data, metadata management and communication comes at high cost or high overhead. The cliché example in our industry would likely be the output from Mass Spec machines which have a history of filling up a single directory for each experiment with potentially many thousands of tiny files a few kilobytes in size.
-
- Millions of files in a complex file and folder hierarchy: For me the cliché scenario is Illumina instrument shares. Out of all the genomics platforms I see most often in consulting work (Illumina, Pacbio, 454, etc.) it’s always the Illumina folders that have the most complexity and files when looking at things from a filesystem view. Pacbio comes a close second due to deep folder nesting requiring complex traversals to probe the filesystem. Illumina ‘wins’ though – it is not uncommon to see an Illumina/ folder with 4+ million files in it representing the output from a single HiSeq-2500 platform.
-
- Large binary and text files. Biologists still create “databases” of DNA and Protein sequences that are either huge flat text files or a combination of huge flat text files with an associated binary index. The cliché example here would be BLAST formatted databases. In the genomics world, Fastq and SAM files are also quite large. For big binary files the common culprits are massive amounts of very large BAM files (BAM is a binary form of SAM) or binary tar.gz type archives of genome, experiments or data mash-ups.
-
- Access Pattern – Long sequential reads: The cliché example here is any sort of bioinformatics analysis such as a BLAST search requiring an input DNA/protein query file to be compared against a very large BLAST database made up of text files and binary indices. The file access pattern is very much concentrated around long sequential reads of big files.
- Access Pattern – Random IO: Too many examples to count. Across the entire spectrum of molecular modeling, structure prediction, computational chemistry, bioinformatics and genomics we have many small algorithms and workflows that create highly variable random IO access patterns and read/write requests.
Data Sources and Producers
The short explanation here is that there are three primary drivers for data acquisition: data generated internally in on-premise or on-campus labs, data acquired from collaborators or partners via shipment of physical media and finally data downloaded via the internet from public or private data providers.
A bit more detail:
-
- Internally Generated: By far the largest producers of internal data are next-gen genomic sequencing platforms. By sheer data volume and number of instruments installed these systems blow away everything else from a data volume perspective. There certainly are other high-scale data internal data producers that surpass NGS in output but they tend to have smaller installation bases and thus are encountered less often. Non-NGS examples include: Confocal Microscopy w/ high-speed cameras, Long-term live cell imaging experiments, medical and experimental imaging (NMR, PET, etc.) systems, 3D imaging and analysis of any kind. Video data is increasing as well – notably in animal facilities where 24×7 video streams of cages or individual animals are being analyzed to programmatically characterize behavior.
-
- Special note about internal data generation: From an IT and facilities perspective it is important to note that historically the lab environments have been neglected or de-prioritized during campus network and LAN upgrade efforts. This becomes a significant problem when terabytes of data are being generated in labs with small network links back to the core datacenter. Storage and data ingest architectures are often modified to deal with poor networking links to remote laboratories. This is starting to change and BioTeam has been directly involved in a number of significant LAN and campus networking enhancements projects specifically aimed at solving this issue. 10Gigabit Ethernet drops are getting closer and closer to the wet lab bench and the use of 40Gigabit Ethernet at the network core or for building-to-building links is increasing steadily.
-
- Partners and Providers (Physical media): A tremendous amount of data is ingested and swapped via shipments of physical media between sites, collaborators and outsourced sequencing and data providers. The most common form factor is 1-2TB SATA disk drives. Issues and concerns regarding physical data ingest are discussed in more detail below.
-
- Partners and Providers (Internet): Petabytes of data are freely available for download and use ranging from full-text scientific literature to huge amounts of genome, DNA and protein datasets. There are valid business and scientific requirements for scientists and researchers to desire access to or local copies of this data. One example: the last time I looked at the 1000Genomes project, the full uncompressed size of their distribution was 464 terabytes! The majority of people we work with are still using standard internet connections and mainstream download methods (HTTP, FTP, RSYNC, etc.) while a smaller number have moved to GridFTP or Aspera based transfer systems. A very small percentage of our clients are connected to Internet2 or other high speed research networks. A small number of our commercial customers have installed secondary internet connections (often cable modems or other services where low cost and high speed is more of a concern than high availability) specifically to handle bulk data movement to and from the internet. Interest is rising in at-scale ingest solutions including “Science DMZs” architected to get around common issues with standard firewalls and intrusion detection systems.
- Partners and Providers (Internet): Petabytes of data are freely available for download and use ranging from full-text scientific literature to huge amounts of genome, DNA and protein datasets. There are valid business and scientific requirements for scientists and researchers to desire access to or local copies of this data. One example: the last time I looked at the 1000Genomes project, the full uncompressed size of their distribution was 464 terabytes! The majority of people we work with are still using standard internet connections and mainstream download methods (HTTP, FTP, RSYNC, etc.) while a smaller number have moved to GridFTP or Aspera based transfer systems. A very small percentage of our clients are connected to Internet2 or other high speed research networks. A small number of our commercial customers have installed secondary internet connections (often cable modems or other services where low cost and high speed is more of a concern than high availability) specifically to handle bulk data movement to and from the internet. Interest is rising in at-scale ingest solutions including “Science DMZs” architected to get around common issues with standard firewalls and intrusion detection systems.
Next-Gen Genomic Sequencing Workflow and Data Examples
Example: 800 Outsourced Genomes
- Company X prepares 800 samples and sends them to Beijing for sequencing by BGI
- Company also decides as a test case to purchase BGI analysis/bioinformatics services on the samples
- Turnaround time: Several months
- BGI physically ships 30 terabytes of data back to Company X in the form of single-disk SATA enclosures
- All 30 TB of data is manually ingested into online storage by a PhD scientist manually copying data
- The 30TB of data breaks down into two main types
- Type 1: BGI analysis results for each sequenced sample. Genomes compared against a reference and differences are noted and distributed via VCF Files
- Type 2: All of the raw “evidence” used to justify the VCF files produced by the bioinformatics analysis. Distributed in the form of BAM and Fastq files. In addition to containing the actual genome for each sample these files also allow Company X to perform their own bioinformatics analysis as well as spot-check the BGI produced VCF calls.
- The analysis result files (VCF format) consume roughly 1GB per submitted sample
End result: For 800 outsourced genomes the data produced breaks down into roughly 29 terabytes of BAM and FASTQ files that will rarely (if ever) be looked at and 1TB of variant calls in the form of VCF files which represent the majority of the desired scientific result data.
Example: Internal NGS Sequencing
- Company X operates a single HiSeq 1000
- Instrument can process 32 genomes in parallel and takes 10 days to produce files that can be moved elsewhere for analysis
- Final instrument data for the 32 samples shows up “all at once” so this is an “every 10 days” operation
- Summary: 32 genomes produced every 10 days per HiSeq 1000 platform (and plans are underway to upgrade the HiSeq to a 2500 …)
- In this example, instrument data is first moved to a local large-RAM Linux server for batch conversion into standard FASTQ file format
- 10GB FASTQ per sample is the end result. Summary: 320GB in new raw data every 10 days per HiSeq 1000
- The 320GB of FASTQ files are processed via a commercial software suite (CLCBio)
- CLCBio is cluster-aware and knows how to submit jobs to the local HPC Linux compute farm
- CLCBio produces 1GB of VCF variant call file data for every 10GB FASTQ input file
End result: Every 10 days 32 samples are processed. The instrument data is moved to a scale-out NAS platform and a large-RAM Linux server is used to bulk convert the Illumina data into 320 gigabytes of FASTQ files. The FASTQ files are then processed via CLCBio and the NAS-attached HPC compute farm in order to generate an additional 32 gigabytes of variant data in the form of VCF files. The 32 gigabytes of VCF data is “kept forever” while the 320 gigabytes FASTQ dataset may be kept for years or months but is more often simply deleted or archived over time.
Example: HiSeq 2000 File sizes and processing space requirements
This is a distilled summary of a conversations with Chris Dwan, a former BioTeam guy and all-around excellent resource.
- BSQ files come off the instrument. 2 bits are needed for the DNA basecall, 8 bits are required to store the related quality values
- This gives a file size requirement of 1.25 bytes of data needed to store each DNA base
- Most people demultiplex the BSQ files into BAM files. The data is still unaligned at this point but file storage requirements still stay at 1.25 byes per base at this point. End result is that the BAM+BSQ filesize requirements are roughly 2x the size of the BSQ data
- It is when people unpack the BAM into FASTQ format that the file size blows up significantly. Much of the file size expansion can be reclaimed by using gzip compression on the FASTQ files. Pipelines and workflows may have to be built that automatically handle GZIP compression and decompression
- FASTQ files are overwhelmingly the largest file type. Variant data is typically 1-2GB per genome sample
- 30x coverage on a HiSeq 2000 sequencing run requires about 130GB of space for BSQ files. This requirement blows up to a working directory requirement of about 2TB for any use case where EVERYTHING needs to be unpacked, accessible and available (BSQ, BAM, FASTQ source, reference source, FASTQ aligned, etc)
- High throughput labs involved in this work appear to store lossily compressed versions of BAM files and pristine variant call data. All of the other files and data can be regenerated from the BAM and variant data in a matter of days if needed
Scientific Data Movement via Physical Ingest


Summary: It is very easy to talk about data movement via copying from physical media and we hear these conversations often. However, when you actually try to do this in the real world with scientific data it turns out to be incredibly resource intensive and also potentially very risky when done casually by scientists working without a SOP and little understanding of checksum and data consistency. Serious ingest of physical media needs serious planning and attention if it is going to be done effectively and safely. There is a non-trivial risk of scientific data loss, corruption or file truncation in environments where physical ingest is handled in a casual manner.
This is probably the most frequent “high risk” activity we regularly see onsite at clients in 2013.
There is a tremendous amount of life science data moving around and between organizations via disk drives and other physical media. People who have not done physical data movement for real in any significant way are often casual and glib about the process (“… no big deal, it’s just copying files … “). This is a hugely mistaken attitude. Below are some thoughts collected over the last few years …
Issues
-
- Productivity loss: I’ve lost count of the number of highly-paid, highly-specialized PhD scientists I’ve encountered who have been saddled with the responsibility for receiving and processing disk drives as they show up in the mail. Some of the most highly paid and highly skilled employees are spending many hours per week/month manually tracking FedEx shipments and copying files onto the network. To me — this is a ludicrous waste of resources.
-
- Network utilization: Network designers rarely assume terabyte-scale data flows coming from desk side or office network drops, in organizations where the access-layer switching is oversubscribed by design there could be performance problems and other unfortunate side effects (VoIP, conferencing etc. )
-
- Little automation: Far too many people doing manual “drag and drop” copying with zero automation or remote progress monitoring. You better hope that the user does not habitually click “OK” too quickly and dismisses notifications regarding transfer and copy errors.
-
- Risky procedures: Organizations that have senior scientists wasting time manually copying files are also most likely to not have an actual protocol or SOP written up to govern the act of physical ingest. The end result is that novice users use manual, ad-hoc or ‘casual’ copying methods to move data off of the physical media. No constancy checking, no MD5 checksum comparisons and no review of the file manifest to confirm that every file made it into the remote share.
Recommendations
-
- Be serious: Physical ingest of scientific data needs careful thought and planning. You’ve worked hard (or paid) for this data and equal care should be taken to ensure that it is ingested efficiently and accurately
-
- Monitor Resources: Data movement can be pretty time consuming, especially if done by hand over standard Ethernet networks. The time and effort required to manually process data off of physical media should be tracked some way simply to ensure that resources and time is being spent effectively. At some point you’ll want to do the “what is the per hour employee cost of this work vs. the monthly cost of a bigger internet connection?“
-
- Dedicated Hardware: The standard corporate desktop may not be an ideal ingest station, most notably they tend to be underpowered from the perspective of having the internal IO, CPU and RAM necessary to properly perform MD5 checksums on each file or some other type of “before and after” data consistency checking. Serious ingest should be done via a dedicated workstation if only for the simple reason of not unnecessarily tying up a desktop that a researcher needs in order to work. Tower-style PC systems are a great choice as they can easily be kitted out with additional hot swap drive bays, USB 3.0 / SATA / eSATA expansion cards as well as (potentially) 10Gigabit network cards.
-
- Thoughtful Location: Make conscious decisions about where ingest workstations reside from a network topology perspective. Copying from a desktop environment at the far-end of an oversubscribed gigabit Ethernet access layer switch may not be the best decision. Consider installing dedicated network cables or using a special VLAN upon which the network engineers can tweak QoS settings to ensure that other business users remain unaffected
-
- Network: Edge or access layer switches feeding desktop environments with 1Gig links may often have 10Gigabit capabilities or open ports that are not fully consumed by trunking or upstream connectivity. A $400 SFP+ module and a $75 cable may be the only thing preventing you from running a 10Gig connection to the ingest workstation. This sort of thing is worth researching with the local networking folks.
-
- Develop methods and procedures: A formal written SOP should be developed and followed to ensure that data ingest occurs properly, successfully and safely
- Do something useful with the physical devices: It’s comical how many bookshelves I’ve seen in researcher offices packed full of processed disk drives. At a minimum these devices should be treated as potential offsite backup or disaster-recovery assets. The simple act of storing the disk drives in a fireproof media cabinet on a different floor or in a different building may negate the need to formally backup or archive the ingested data.
My ideal high-scale physical ingest setup: Drives get shipped directly to the datacenter or IT operations desk where a specialized ingest server has been installed with direct connections into the primary scientific storage tier. IT operations staff manage the data ingest, carefully following a written SOP and notifying the scientific data owner when the process is initiated and completed. The ingested drive is then moved offsite or into a fireproof media cabinet as an inexpensive DR asset.
Scientific Data Movement via Network Ingest
After being involved in a small number of data-movement and ingest projects I tend to favor network-based movement, even if it requires investment in hardware, software or a faster internet circuit. These costs tend to be cheaper than human/operational burden (and risks!) of running a large physical ingest process. In addition – some investments, such as a larger pipe to the internet will benefit the entire organization.

It’s amazing what a Gigabit Ethernet pipe to the internet will allow you to do. They may be more affordable than you think (or you’ve been told ...). It is worth exploring what the actual cost would be to increase onsite internet connectivity. The cost of a fatter pipe will quite often be cheaper than the human and operational cost of doing lots of manual data ingest and movement.
Sometimes it can be quite hard to figure out the actual cost of a “bigger internet circuit”, especially in large enterprise organizations where the Research IT people may not have strong contacts among the Telecom and WAN groups. If you just ask the question casually, the Official Answer may be calculated based on current practices and WAN circuits that are business-critical and carry high-level SLA agreements. This is why the WAN engineers at a company running a complex MPLS ring as it’s primary communication infrastructure may tell scientists that “… what you are asking for will cost us between $10,000 and $30,000 a month.” – a high price that would scare off many Bio-IT people. That may represent an honest answer but it may not represent the only answer.
Fast internet pipes do not require exotic and expensive telecommunication circuits. They may not require bulletproof guarantees and SLAs. They may not have to be deployed everywhere. With appropriate buy-in from the networking people and support from senior management it may be possible to get a very fast pipe at a reasonable cost.
-
- A small pharmaceutical company has installed a cable modem traditionally marketed to small business and home office professionals directly into their machine room. This cable modem is used for downloading massive public domain data sets at high speed. The device is dirt cheap relative to their carrier-grade primary circuits, works well and keeps a ton of internet traffic off of the expensive and slower link
-
- A large pharmaceutical company with a big MPLS ring and WAN circuits that span the globe has recently installed a 1 Gigabit Ethernet internet circuit into a datacenter where most of the scientific computing data and HPC resources reside. This single circuit is very reasonably priced and the pharmaceutical company has used inexpensive routing, firewall and proxy appliances to expose this fast internet pipe only to the scientific computing and HPC environment. This circuit is also likely to be the location where hardware VPN appliances are installed if they ever plan to make a persistent link into an Amazon VPC environment.
Network Ingest Recommendations
-
- Tune your tools: Copying or replicating data via Unix tools like FTP, rsync or rsync-over-ssh is fine but anyone using these methods should take a careful dive through the wonderful ESNET FasterData website hosted over at http://fasterdata.es.net/ — in particular for SSH/SCP users please pay careful attention to the “Say no to SCP” write-up
-
- Cloud xfer tools: There are a number of open source and commercial tools for easy cloud data movement or folder synchronization. Sometimes these tools default to the “easy” mode of operation and do not enable by default behavior that you may want. Spend some time reading the documentation and diving into the configuration and preferences interfaces. For Amazon in particular you want to make sure you have enabled the features that trigger the checksum comparisons, tuned for the proper number of HTTP threads and enabled multipart HTTP uploads and downloads. If you have a storage product that does cloud replication or provides cloud gateway services the suggestion remains the same — dive into the config details and make sure that the various data integrity and parallel operation features are enabled and configured properly.
-
- Tools for serious data movement: Our old glib advice about specialized data movement tools was “go to the supercomputing conference and check out the code used by the group that wins the annual bandwidth challenge contest“. That is no longer really necessary any more as the ‘next-gen’ toolsets have largely settled down in the Bio-IT space. If you go out to conferences and meetings and talk to people who do hardcore network data movement you will find that our community has basically consolidated around two software packages: the freely available GridFTP implementation and related commercial data portal services from GlobusOnline or the commercial software, library, plugin and portal suite from Aspera.
-
- Beyond 10Gig: If high speed research networks, 40Gig or 100Gig networking is in your future, start sitting down with your risk assessment, network security and firewall people now. You will quickly find that the traditional methods for network security including stateful firewalls, deep packet inspection and application-layer intrusion detection do not easily scale past the 1Gigabit mark. You really don’t want to know what a firewall capable of doing DPI on a 10Gigabit optical network segment will cost! The good news is that high-speed movement of research data is a problem that many fields have faced in the past and there is an emerging collection of successful design patterns and reference architectures. The bad news is that these new patterns and methods may be out of the comfort-zone of your standard corporate firewall administrator or security engineer. The best starting point for experts and novice alike is the http://fasterdata.es.net/science-dmz/ site which has collected many of the best practices around what is now commonly referred to as “Science DMZ”.
Cloud Storage
One storage ‘meta’ issue
It is important to comprehend the core ‘meta’ issue affecting storage and data locality before diving into all of the interesting economic and capability factors of cloud-resident block or object storage. What I’ve learned over time across many projects is that scientific computing power is in many cases just a straightforward commodity that is easy to acquire and deploy wherever and whenever one needs it.
This actually has a tangible and real world effect on the look and feel of modern informatics IT infrastructure — the simple fact is that users, applications and computing will naturally flow to where the data density is highest. Given the relative “weight” of data under management it is almost always easier to bring the users and compute to wherever the data resides.
This is why pharma and biotech companies are shrinking “islands” of HPC installed at satellite facilities and instead are choosing to consolidate scientific computing within one or two primary datacenters. Operationally and technically it is far easier to centralize the storage(especially petabyte-scale installations) and compute and bring the users in over the network. Even the edge cases like latency sensitive modeling and 3D visualization requirements are falling away as VPN or WAN-friendly graphical desktop clients get better at 3D and GPU-accelerated visualization.
The “storage as center of gravity” viewpoint should influence how the cloud is thought of as a potential storage target.
-
- For some people, this may mean that it makes sense to invest in capabilities that enhance the ability to rapidly fire up analytical capability in the cloud so that it can compute directly against data that is already nearby
-
- For others, this may mean an internal stance centered around keeping the primary scientific data tier on-premise with cloud storage only used for advantageous use cases like sharing data with collaborators or as a deep archive or disaster recovery asset
Recommendations
-
- Plan for it: Using or accessing cloud-resident storage is something you WILL be doing and this assumption should be part of multi-year research IT infrastructure planning. If not for capability or economic reasons than because a collaborator or vendor is using the cloud as a data distribution point.
-
- Start the ‘slow’ stuff now: Accessing the cloud from a technology standpoint is very easy. Lawyers, policies, procedures and risk assessment programs can take much longer. I’ve personally seen cloud pilot projects where the baseline functionality could be built in a day or two get stalled for months or even years once the lawyers and risk management people got involved. If you think your organization moves very slowly when it comes to new methods and techniques than start the process moving sooner rather than later so that all of the process and policy stuff is complete or almost complete by the time you want to do something useful.
-
- The cloud will be a storage tier option: Many of our storage products are already running 64 bit Unix systems under the hood on powerful x86_64 CPUs and more than a few of them can run apps or even hypervisors natively. When you combine the abilities of a powerful “Unix-like” OS running on top of local storage with the fact that most cloud object storage services are a simple RESTful HTTP call away it becomes very very obvious that modern storage gear should easily be able to treat remote object stores as nothing more than a distant (and high-latency) disk store. Avere does this now with their recent announcement and you should expect this to be a feature in more and more storage products over time.
Politics and IT Empires: Dealing with cost FUD
Cloud economics is a complicated area. Right now, if you have a particular “pro-cloud” or “anti-cloud” agenda it is fairly easy to generate cost-based arguments that favor whatever view you are attempting to promote within your organization. It is, however, getting harder to fog up arguments surrounding the cost of cloud storage. The economics are getting harder to spin as services like AWS S3 continuously announce price cuts and volume discounts (AWS S3 has announced 24 consecutive price reductions through November 2012).
The simple fact is that entities that have petabytes-to-exabytes of spinning disk installed all over the world can leverage economies of scale that non of us regular people can match. This allows some of those entities to sell people like us access to remote storage at a price point that we would find hard to match internally.
The cheesiest arguments are when internal IT people compare the per-terabyte or per-gigabyte cost of a single on-premise storage platform with something like Amazon S3 – conveniently assuming that buildings, electricity, cooling, network, maintenance, upgrades and human staffing are infinite and free.
If you see this happening within your organization it’s worth diving a bit deeper. Replying with a question like this can often prove illuminating:
“… The Amazon S3 control backplane will not even acknowledge a successful PUT operation until the file has landed in three geographically separated datacenters. Can you please redo the cost analysis to reveal the per-gigabyte internal cost of synchronous storage replication between three remote company locations and make sure the estimate includes facility, electrical, cooling, networking, staffing and emergency power overhead costs?“
If you want to compare Amazon S3 with “anything else” then make sure that the comparison is fair. If the alternative does not include synchronous multi-site replication then at least disclose that clearly somewhere within the analysis.
Conversely, I’ve also seen 100% legit cloud storage cost analysis documents that show the cloud to be economically unviable, particularly for scientific computing use cases where large volumes of scientific data would be flowing bidirectionally into and out of the cloud provider on a continuous basis (IaaS providers may charge volume-based transfer fees data flows entering or exiting the provider).
What this boils down to is that when it comes to the cost of cloud computing, Bio-IT people should understand that this is an area where politics can fog up the picture. People should generally take a “trust but verify” stance with all of the information provided by both insiders and outsiders.
Security and Cloud Storage
Security is a big deal. Especially now that we know our sovereign governments are often the ones stealing the private keys, tapping the circuits and working to weaken and sabotage critical industry encryption standards. I won’t pretend to be a security expert but I can share some opinions and anecdotes from real work …
-
- The answer to most security concerns is strong encryption. Don’t trust your cloud provider? Don’t trust your internet links? Encrypt every byte of information before it leaves your premise and keep the keys to yourself.
-
- I’ve found that the technical act of encryption and decryption is pretty straightforward. Properly managing your private key infrastructure, exchanging keys and handling key rotation and revocation issues is the hard and operationally nasty part. The simple truth is that encryption is easy, properly managing a security and key management infrastructure can be very difficult. This needs to be understood by all before any serious data encryption effort is undertaken.
-
- My personal FUD detectors get lit up when I see midlevel IT professionals demanding that a cloud vendor have certain security features or 3rd party certifications that they themselves have not bothered to implement internally. If security requirement X and third party audit certification Y is so critical then why has your organization failed to obtain them?
-
- Have you seen the Amazon CloudTrail announcement? Full-on audit logs for AWS API activity. This is going to make a lot of security and compliance people happy.
-
- It is relatively easy to get deeply technical and straight security answers out of the major cloud providers. I’ve seen this with every cloud provider I’ve worked with. All of the major players understand that security is a crucial concern amongst potential customers and all of them bend over backwards to make incredibly skilled and highly technical experts available for deep conversations with your security engineers and risk management professionals. The blunt truth is that you may not LIKE what they tell you but you WILL get honest technical data and straightforward answers from highly capable people. Initial contacts with a new provider might involve a layer or two of basic pre-sales people but those folks will know how to get the heavy hitters on the phone when needed.
-
- I personally do not have a lot of fear about doing real work on a platform belonging to a major provider, especially when I compare them against the real world IT environments and staff practices I see almost every day. Microsoft, Google, Amazon etc. all have many years of experience running incredibly massive internet-facing systems operating in an insanely hostile networking environment. I generally feel that these providers run tighter shops with more stringent operational and security controls and that their methods are audited, tested and updated more frequently than the IT environments of the typical life science company. Others may disagree.
Wrap up & Revision History
December 2013: This section is for tracking edits, updates and corrections over the lifespan of this document.


